3 Intro to the Tidyverse
https://learn.datacamp.com/courses/introduction-to-the-tidyverse
Main functions and concepts covered in this chapter:
- pipes (
%>%) filter()arrange()mutate()- scatterplots (
geom_point()) - graph aesthetics (color, size, etc.)
facet_wrap()summarize()group_by()- line plots (
geom_line()) - bar plots (
geom_col()) - histograms (
geom_histogram()) - boxplots (
geom_boxplot())
Packages used in this chapter:
## Load all packages used in this chapter
library(tidyverse) #includes dplyr, ggplot2, and other common packages, so we typically just load tidyverse instead of loading them separately (otherwise people tend to load the the same packages multiple times which slows things down)## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
## ✔ ggplot2 3.4.0 ✔ purrr 0.3.5
## ✔ tibble 3.1.8 ✔ dplyr 1.0.10
## ✔ tidyr 1.2.1 ✔ stringr 1.5.0
## ✔ readr 2.1.3 ✔ forcats 0.5.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()library(gapminder)
library(Lock5Data)Datasets used in this chapter:
## Load datasets used in this chapter
# We use gapminder package
# it is loaded automatically when we load the gapminder package
# (i.e., when we call library(gapminder))
# We also use the SleepStudy and HappyPlanetIndex dataset
# it is load automatically when we load the Lock5Data package3.1 Data wrangling
Before we get started, it’s a good idea to have a general sense of what’s in the dataset we’ll be using:
str(gapminder)## tibble [1,704 × 6] (S3: tbl_df/tbl/data.frame)
## $ country : Factor w/ 142 levels "Afghanistan",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ continent: Factor w/ 5 levels "Africa","Americas",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ year : int [1:1704] 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 ...
## $ lifeExp : num [1:1704] 28.8 30.3 32 34 36.1 ...
## $ pop : int [1:1704] 8425333 9240934 10267083 11537966 13079460 14880372 12881816 13867957 16317921 22227415 ...
## $ gdpPercap: num [1:1704] 779 821 853 836 740 ...summary(gapminder)## country continent year lifeExp
## Afghanistan: 12 Africa :624 Min. :1952 Min. :23.60
## Albania : 12 Americas:300 1st Qu.:1966 1st Qu.:48.20
## Algeria : 12 Asia :396 Median :1980 Median :60.71
## Angola : 12 Europe :360 Mean :1980 Mean :59.47
## Argentina : 12 Oceania : 24 3rd Qu.:1993 3rd Qu.:70.85
## Australia : 12 Max. :2007 Max. :82.60
## (Other) :1632
## pop gdpPercap
## Min. :6.001e+04 Min. : 241.2
## 1st Qu.:2.794e+06 1st Qu.: 1202.1
## Median :7.024e+06 Median : 3531.8
## Mean :2.960e+07 Mean : 7215.3
## 3rd Qu.:1.959e+07 3rd Qu.: 9325.5
## Max. :1.319e+09 Max. :113523.1
## # To see how many observations per year, we can convert year to a factor and summarize it
summary(as.factor(gapminder$year))## 1952 1957 1962 1967 1972 1977 1982 1987 1992 1997 2002 2007
## 142 142 142 142 142 142 142 142 142 142 142 1423.1.1 Pipe Operator
%>%: the pipe operator: take whatever is before it, and feed it into the next step.
In other words, a %>% sum(b) is the same as `sum(a+b)
a <- 1
b <- 2
a %>% sum(b)## [1] 33.1.2 Filter
In the tidyverse (dplyr is one of the tidyverse packages), functions that do things to or with a dataset are called “verbs”. Our first verb is filter().
The filter() verb extracts particular observations based on a condition
For example, we can get only observations for year 1957
# Filter the gapminder dataset for the year 1957
gapminder %>% filter(year == 1957)## # A tibble: 142 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1957 30.3 9240934 821.
## 2 Albania Europe 1957 59.3 1476505 1942.
## 3 Algeria Africa 1957 45.7 10270856 3014.
## 4 Angola Africa 1957 32.0 4561361 3828.
## 5 Argentina Americas 1957 64.4 19610538 6857.
## 6 Australia Oceania 1957 70.3 9712569 10950.
## 7 Austria Europe 1957 67.5 6965860 8843.
## 8 Bahrain Asia 1957 53.8 138655 11636.
## 9 Bangladesh Asia 1957 39.3 51365468 662.
## 10 Belgium Europe 1957 69.2 8989111 9715.
## # … with 132 more rowsWe see 10 rows, and it says there are 132 more rows. We know from the summary above that there are 142 observations per year, so that makes sense.
You can also filter for multiple conditions:
# Filter for China in 2002
gapminder %>% filter(year==2002 & country == "China")## # A tibble: 1 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 China Asia 2002 72.0 1280400000 3119.# Filter for China or Japan in 2002
gapminder %>% filter(year==2002 & (country == "China" | country == "Japan"))## # A tibble: 2 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 China Asia 2002 72.0 1280400000 3119.
## 2 Japan Asia 2002 82 127065841 28605.# China since 1990
gapminder %>% filter(year>=1990 & country == "China")## # A tibble: 4 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 China Asia 1992 68.7 1164970000 1656.
## 2 China Asia 1997 70.4 1230075000 2289.
## 3 China Asia 2002 72.0 1280400000 3119.
## 4 China Asia 2007 73.0 1318683096 4959.3.1.3 Arrange
You use arrange() to sort observations in ascending or descending order of a particular variable. Use desc() around the variable name to sort in descending order (default is ascending).
# Sort in ascending order of lifeExp
gapminder %>% arrange(lifeExp)## # A tibble: 1,704 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Rwanda Africa 1992 23.6 7290203 737.
## 2 Afghanistan Asia 1952 28.8 8425333 779.
## 3 Gambia Africa 1952 30 284320 485.
## 4 Angola Africa 1952 30.0 4232095 3521.
## 5 Sierra Leone Africa 1952 30.3 2143249 880.
## 6 Afghanistan Asia 1957 30.3 9240934 821.
## 7 Cambodia Asia 1977 31.2 6978607 525.
## 8 Mozambique Africa 1952 31.3 6446316 469.
## 9 Sierra Leone Africa 1957 31.6 2295678 1004.
## 10 Burkina Faso Africa 1952 32.0 4469979 543.
## # … with 1,694 more rows# Sort in descending order of lifeExp
gapminder %>% arrange(desc(lifeExp))## # A tibble: 1,704 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Japan Asia 2002 82 127065841 28605.
## 4 Iceland Europe 2007 81.8 301931 36181.
## 5 Switzerland Europe 2007 81.7 7554661 37506.
## 6 Hong Kong, China Asia 2002 81.5 6762476 30209.
## 7 Australia Oceania 2007 81.2 20434176 34435.
## 8 Spain Europe 2007 80.9 40448191 28821.
## 9 Sweden Europe 2007 80.9 9031088 33860.
## 10 Israel Asia 2007 80.7 6426679 25523.
## # … with 1,694 more rows3.1.4 Mutate
The mutate() verb changes the values in a column or adds a new column.
Change life expectancy from being measured in years to being measured in months
# Use mutate to create a new column called lifeExpMonths
gapminder %>% mutate(lifeExpMonths = lifeExp * 12)## # A tibble: 1,704 × 7
## country continent year lifeExp pop gdpPercap lifeExpMonths
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. 346.
## 2 Afghanistan Asia 1957 30.3 9240934 821. 364.
## 3 Afghanistan Asia 1962 32.0 10267083 853. 384.
## 4 Afghanistan Asia 1967 34.0 11537966 836. 408.
## 5 Afghanistan Asia 1972 36.1 13079460 740. 433.
## 6 Afghanistan Asia 1977 38.4 14880372 786. 461.
## 7 Afghanistan Asia 1982 39.9 12881816 978. 478.
## 8 Afghanistan Asia 1987 40.8 13867957 852. 490.
## 9 Afghanistan Asia 1992 41.7 16317921 649. 500.
## 10 Afghanistan Asia 1997 41.8 22227415 635. 501.
## # … with 1,694 more rows3.1.5 Combining verbs
We can also combine multiple verbs. Each verb produces output. Using pipes, we just pipe the output of one step into the next verb.
# Sort countries by population in the year 2007
gapminder %>%
filter(year==2007) %>%
arrange(desc(pop))## # A tibble: 142 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 China Asia 2007 73.0 1318683096 4959.
## 2 India Asia 2007 64.7 1110396331 2452.
## 3 United States Americas 2007 78.2 301139947 42952.
## 4 Indonesia Asia 2007 70.6 223547000 3541.
## 5 Brazil Americas 2007 72.4 190010647 9066.
## 6 Pakistan Asia 2007 65.5 169270617 2606.
## 7 Bangladesh Asia 2007 64.1 150448339 1391.
## 8 Nigeria Africa 2007 46.9 135031164 2014.
## 9 Japan Asia 2007 82.6 127467972 31656.
## 10 Mexico Americas 2007 76.2 108700891 11978.
## # … with 132 more rowsNote how the pipe goes at the end of a line when we want to use another verb
We can also use variables created in earlier steps. Here we create the life expectancy in months variable, and then sort by it (after filtering to only have 2007)
# Filter, mutate, and arrange the gapminder dataset
gapminder %>% filter(year == 2007) %>%
mutate(lifeExpMonths = 12 * lifeExp) %>%
arrange(desc(lifeExpMonths))## # A tibble: 142 × 7
## country continent year lifeExp pop gdpPercap lifeExpMonths
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656. 991.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725. 986.
## 3 Iceland Europe 2007 81.8 301931 36181. 981.
## 4 Switzerland Europe 2007 81.7 7554661 37506. 980.
## 5 Australia Oceania 2007 81.2 20434176 34435. 975.
## 6 Spain Europe 2007 80.9 40448191 28821. 971.
## 7 Sweden Europe 2007 80.9 9031088 33860. 971.
## 8 Israel Asia 2007 80.7 6426679 25523. 969.
## 9 France Europe 2007 80.7 61083916 30470. 968.
## 10 Canada Americas 2007 80.7 33390141 36319. 968.
## # … with 132 more rows3.1.6 Creating new dataset from old dataset
Sometimes we want to use a dataset a lot after we’ve applied verbs to the original dataset. For example, in the next DC chapter on data visualization, they have us create a dataset with just 1952. To do this, just save the output into a new variable.
gapminder_1952 <- gapminder %>%
filter(year == 1952)3.2 Data visualization
#Using Sleep Study data set to create new data set with 7 out of the original 23 variables.
sleepstudy <- SleepStudy %>%
select(Gender, ClassYear, GPA, AnxietyScore, Happiness, AlcoholUse, AverageSleep)
#Changing class year from integer to categorical variable.
sleepstudy$classyear <- as.factor(ifelse(sleepstudy$ClassYear < 2, 'First-Year', ifelse(sleepstudy$ClassYear < 3, 'Sophomore', ifelse(sleepstudy$ClassYear < 4, 'Junior', 'Senior'))))
#Changing gender from integer to categorical variable.
sleepstudy$gender <- as.factor(ifelse(sleepstudy$Gender < 1, "Female", "Male"))
str(sleepstudy)## 'data.frame': 253 obs. of 9 variables:
## $ Gender : int 0 0 0 0 0 1 1 0 0 0 ...
## $ ClassYear : int 4 4 4 1 4 4 2 2 1 4 ...
## $ GPA : num 3.6 3.24 2.97 3.76 3.2 3.5 3.35 3 4 2.9 ...
## $ AnxietyScore: int 3 0 18 4 25 8 0 2 16 11 ...
## $ Happiness : int 28 25 17 32 15 22 25 29 29 30 ...
## $ AlcoholUse : Factor w/ 4 levels "Abstain","Heavy",..: 4 4 3 3 4 1 4 3 3 4 ...
## $ AverageSleep: num 7.18 6.93 5.02 6.9 6.35 9.04 7.52 9.01 8.54 6.68 ...
## $ classyear : Factor w/ 4 levels "First-Year","Junior",..: 3 3 3 1 3 3 4 4 1 3 ...
## $ gender : Factor w/ 2 levels "Female","Male": 1 1 1 1 1 2 2 1 1 1 ...3.2.1 Scatterplots
#Creating a scatter plot that compares GPA to the average amount of hours slept per night.
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep)) +
geom_point() +
xlab("GPA") + ylab("Avevage Hours of Sleep")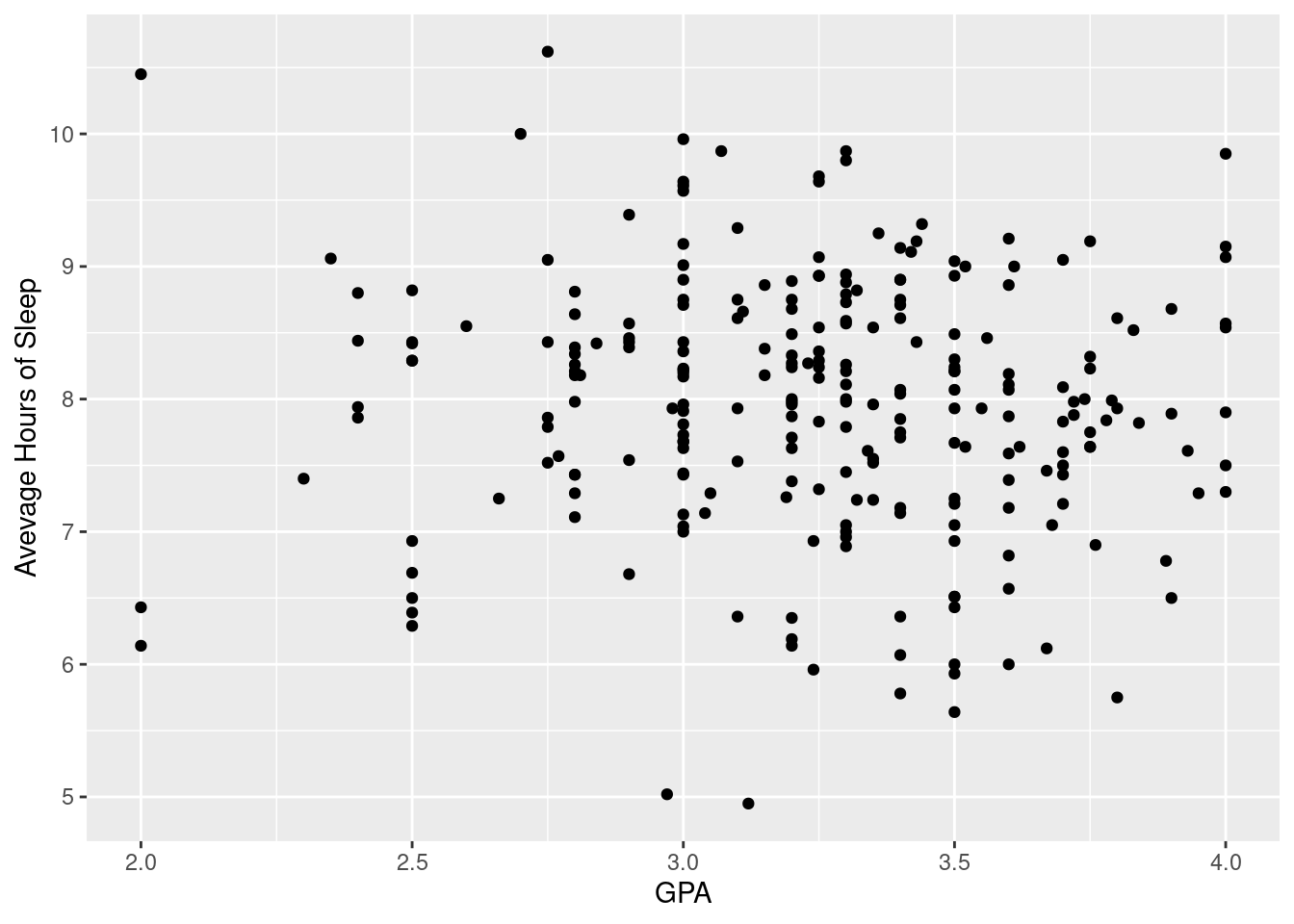
3.2.2 Log Scaling
When a variable is spread over several orders of magnitude, it’s a good idea to put the variable on a log scale in order to spread out the data points more. This makes it easier to see the correlation between the variables. scale_y_log(10).
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep)) +
geom_point() + scale_x_log10() +
xlab("GPA") + ylab("Avevage Hours of Sleep")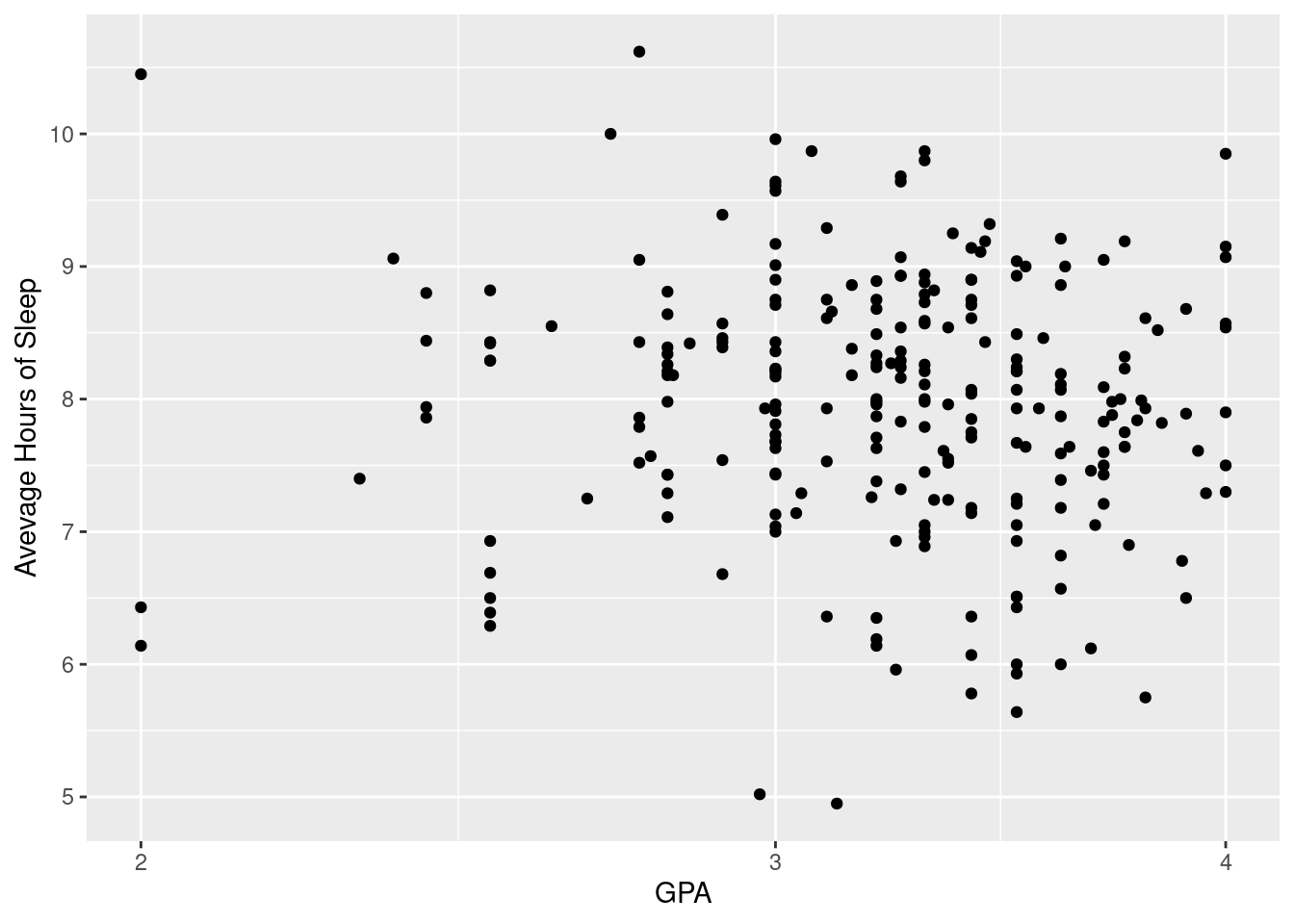
3.2.3 Graph Aesthetics
#Using the same graph as above, we add a third variable, class year, as color. Color is good for categorical variables, though it can be used for continuous variables.
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep, color = classyear)) +
geom_point() +
xlab("GPA") + ylab("Average Hours of Sleep")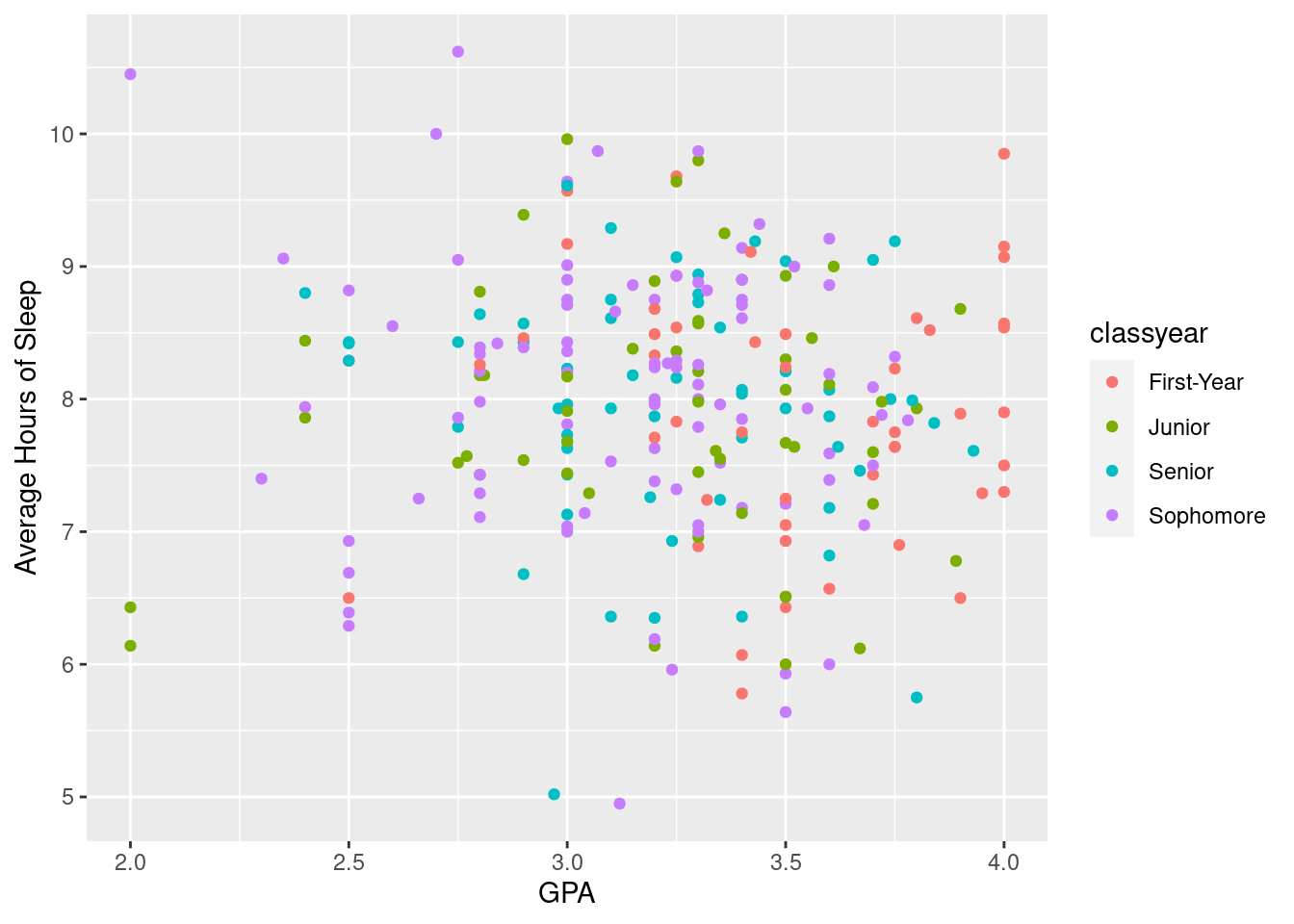
#Using the same graph as above, we add a fourth variable, anxiety score, as size. Here, anxiety score is a numerical variable and the varying sizes can show how high or low the anxiety score is.
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep, color = classyear, size = AnxietyScore)) +
geom_point() +
xlab("GPA") + ylab("Average Hours of Sleep")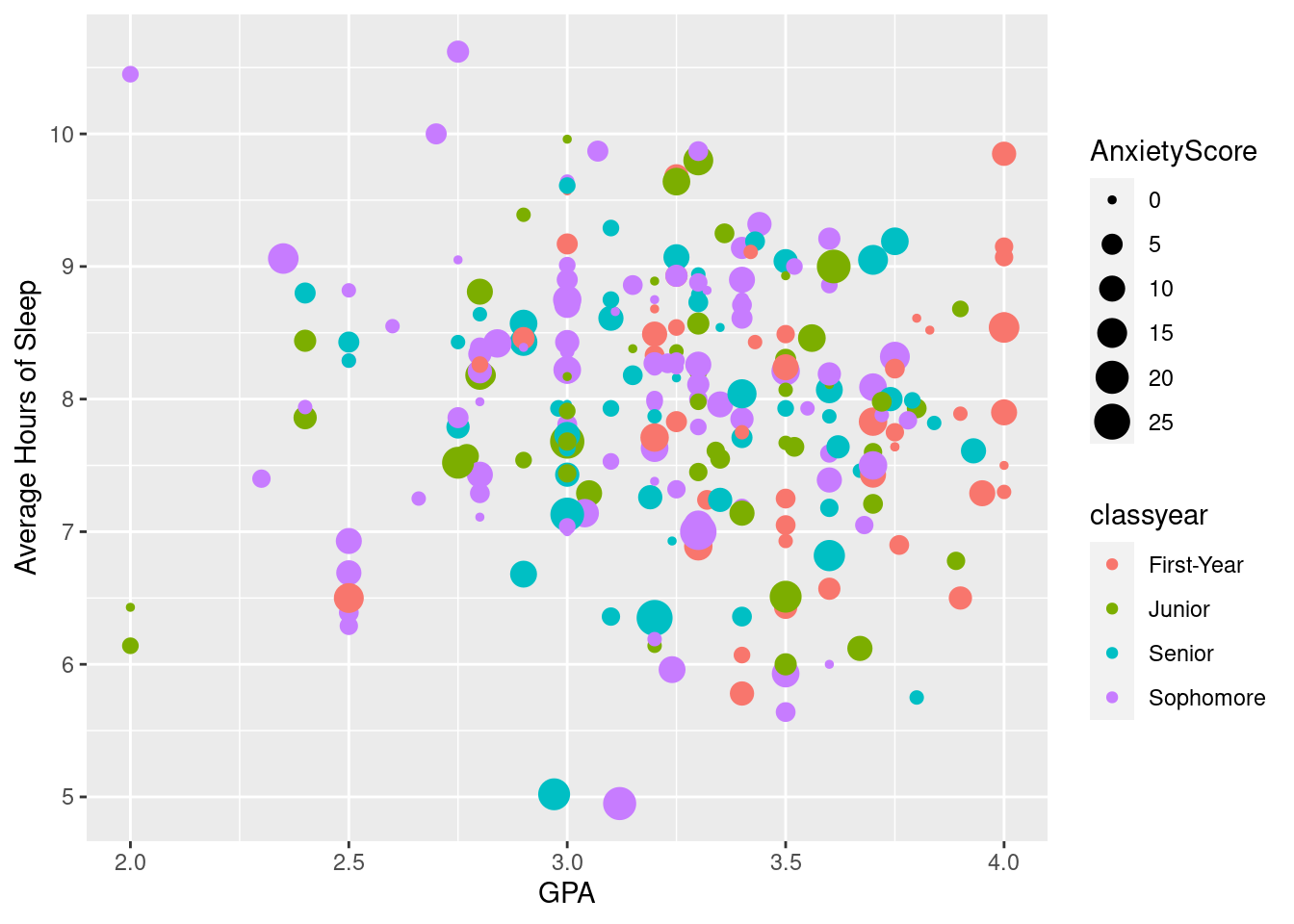
3.2.4 Faceting
facet_wrap()) is a great way to break up your graph into subplots. The variable used to divide the graph should be discrete, though, as a continuous variable would produce far too many subplots. Faceting allows you to look at trends specific to one variable. For example, we can look at how GPA is related to sleep, specific to class year. There may be different relationships between GPA and sleep for sophomores compared to juniors. Faceting can elucidate these differences.
#Here, we create a scatter plot comparing GPA to average hours of sleep and break the graph into subplots based on class year.
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep)) +
facet_wrap(~ classyear) +
geom_point()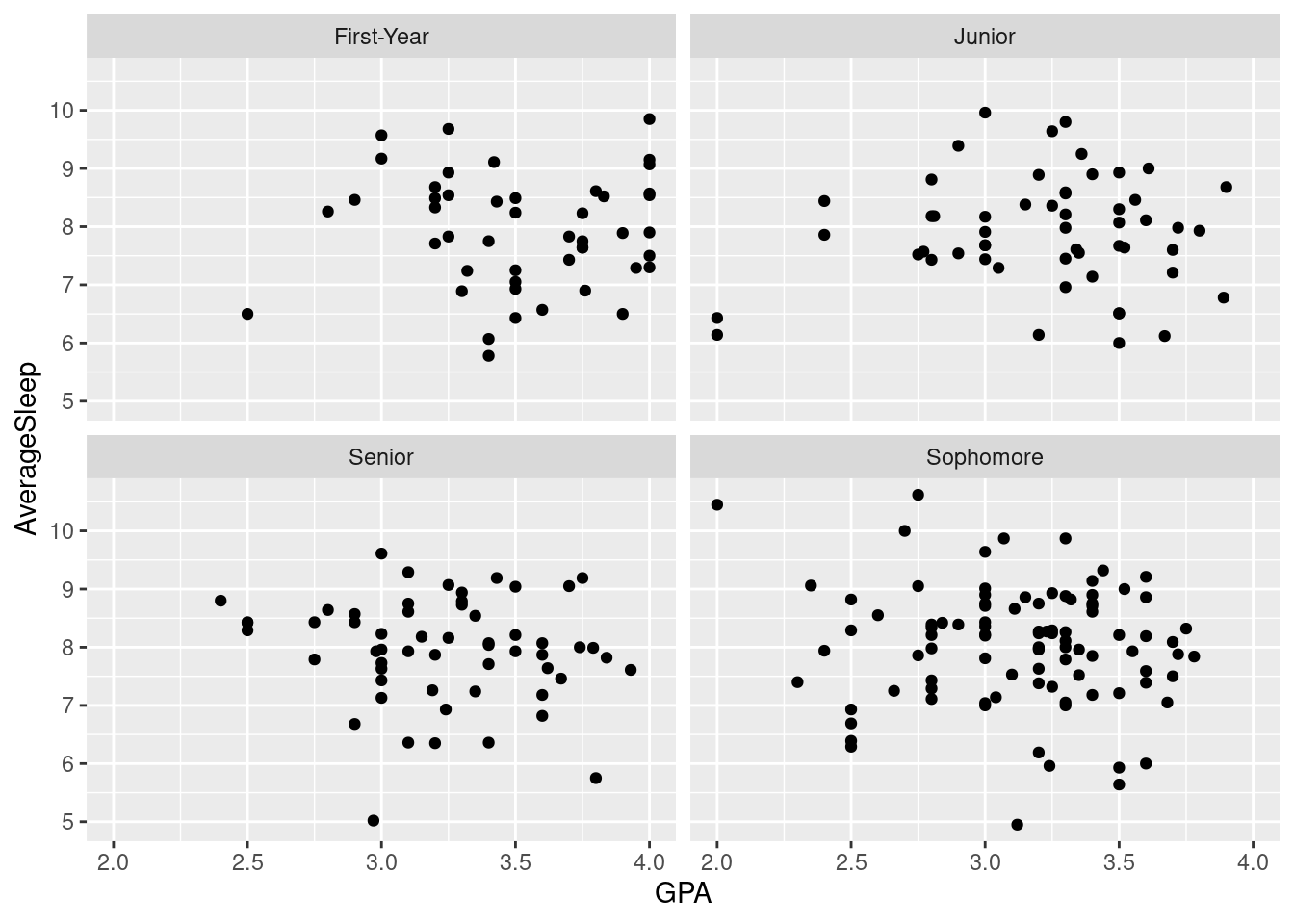
ggplot(sleepstudy, aes(x = GPA, y = AverageSleep, color = gender, size = AnxietyScore)) +
geom_point() +
facet_wrap(~ classyear) 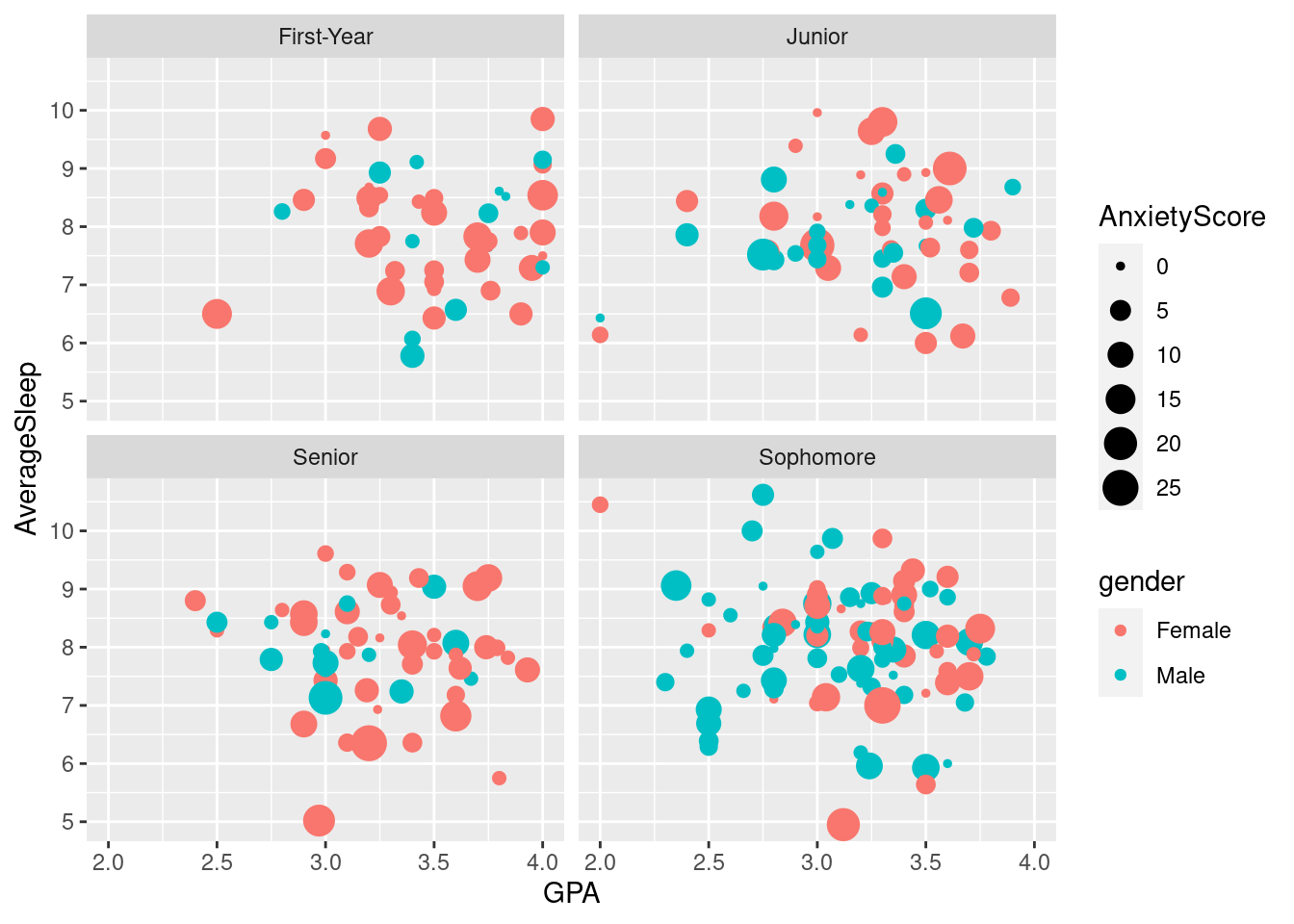
3.3 Grouping and summarizing
3.3.1 Summarizing
summarize() function can be used to visualize information about a certain variable and store it into a single value or vector. For example, we can create a new vector that contains the average amount of sleep across the entire data set. The summarize function is more helpful in conjunction with other functions as seen in examples below.
#Finding the average amount of sleep across the entire data set.
sleepstudy %>%
summarize(meansleep = mean(AverageSleep))## meansleep
## 1 7.965929#Filtering the data for only seniors and then summarizing to see the average amount of sleep for all seniors.
sleepstudy %>%
filter(classyear == "Senior") %>%
summarize(meansleep = mean(AverageSleep))## meansleep
## 1 7.95#We can also summarize for two (or more) different variables at the same time.
sleepstudy %>%
summarize(meansleep = mean(AverageSleep), maxhappy = max(Happiness))## meansleep maxhappy
## 1 7.965929 353.3.2 Grouping
group_by() function groups a data set. It is not especially helpful and will not produce any output unless used with another function, such as summarize(). Using group_by() with summarize() can visualize different metrics of a variable for each category of the variable used to group the data set. We group the data set by a discrete variable (usually categorical but not always) and use summarize() to find, for example, average sleep time for each category in the grouping variable.
#Here we group the data by class year and summarize the data to find the average sleep time and anxiety score for each class year.
sleepstudy %>%
group_by(classyear) %>%
summarize(meansleep = mean(AverageSleep), meananxiety = mean(AnxietyScore))## # A tibble: 4 × 3
## classyear meansleep meananxiety
## <fct> <dbl> <dbl>
## 1 First-Year 7.93 5.09
## 2 Junior 7.90 5.37
## 3 Senior 7.95 5.74
## 4 Sophomore 8.03 5.29sleepstudy %>%
filter(gender == "Female") %>%
group_by(classyear) %>%
summarize(meansleep = mean(AverageSleep), meananxiety = mean(AnxietyScore))## # A tibble: 4 × 3
## classyear meansleep meananxiety
## <fct> <dbl> <dbl>
## 1 First-Year 7.96 6.03
## 2 Junior 7.94 5.81
## 3 Senior 7.93 5.86
## 4 Sophomore 8.13 6.02#We can also group by more than variable. The output breaks down the mean sleep and anxiety score by both class year and gender.
sleepstudy %>%
group_by(classyear, gender) %>%
summarize(meansleep = mean(AverageSleep), meananxiety = mean(AnxietyScore))## `summarise()` has grouped output by 'classyear'. You can override using the
## `.groups` argument.## # A tibble: 8 × 4
## # Groups: classyear [4]
## classyear gender meansleep meananxiety
## <fct> <fct> <dbl> <dbl>
## 1 First-Year Female 7.96 6.03
## 2 First-Year Male 7.84 2.62
## 3 Junior Female 7.94 5.81
## 4 Junior Male 7.84 4.73
## 5 Senior Female 7.93 5.86
## 6 Senior Male 8.01 5.4
## 7 Sophomore Female 8.13 6.02
## 8 Sophomore Male 7.95 4.693.3.3 Grouping Visualizations
#We group the data by anxiety score and find the average sleep time for each anxiety score, saving this to a new data set called "by_anxiety."
by_anxiety <- sleepstudy %>%
group_by(AnxietyScore) %>%
summarize(meansleep = mean(AverageSleep))
#We now graph "by_anxiety" in a scatterplot.
ggplot(by_anxiety, aes(x = AnxietyScore, y = meansleep)) +
geom_point()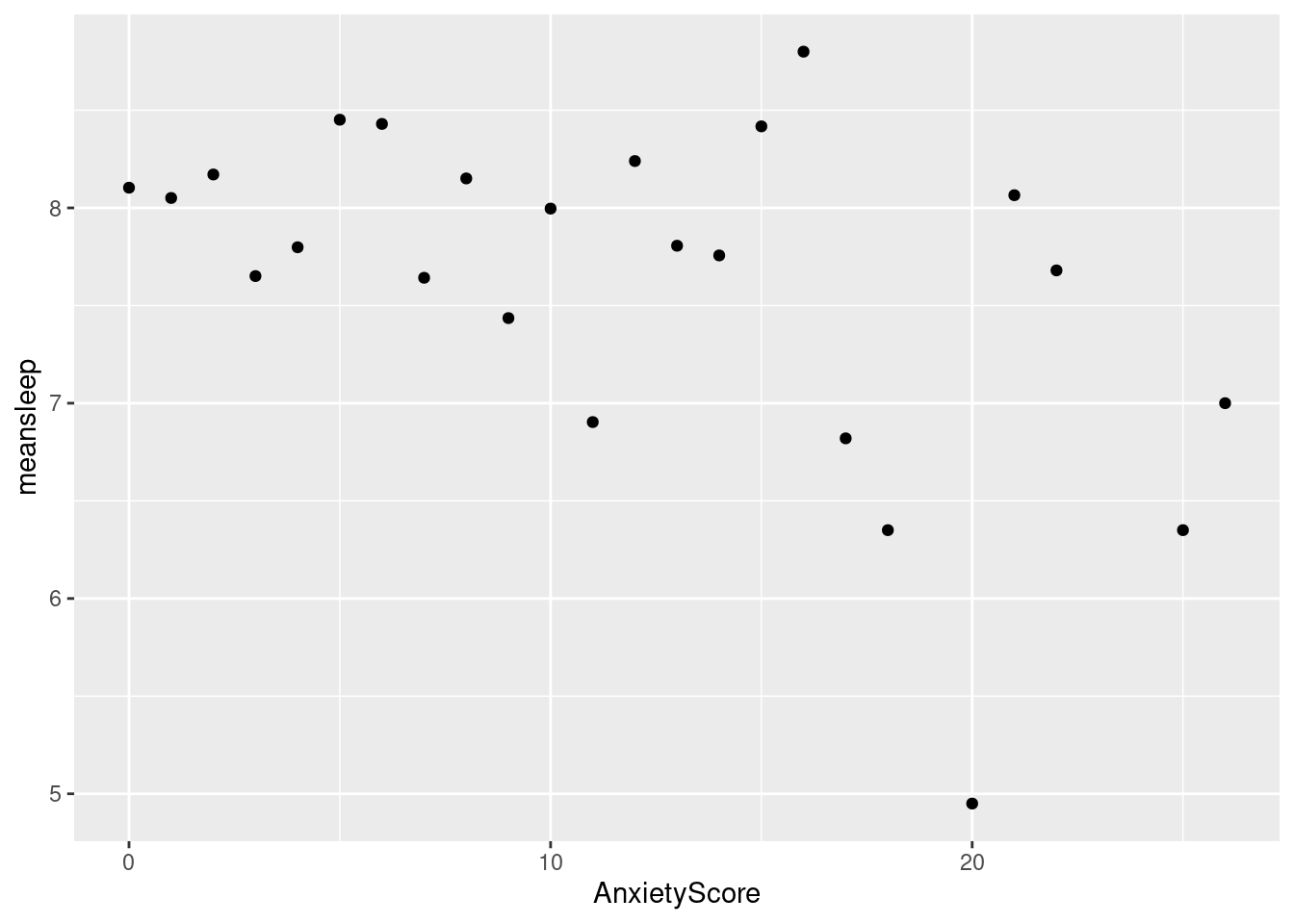
#This graph is similar to the one above but we have filtered the data to only include males.
by_anxiety_male <- sleepstudy %>%
filter(gender == "Male") %>%
group_by(AnxietyScore) %>%
summarize(meansleep = mean(AverageSleep))
ggplot(by_anxiety_male, aes(x = AnxietyScore, y = meansleep)) +
geom_point()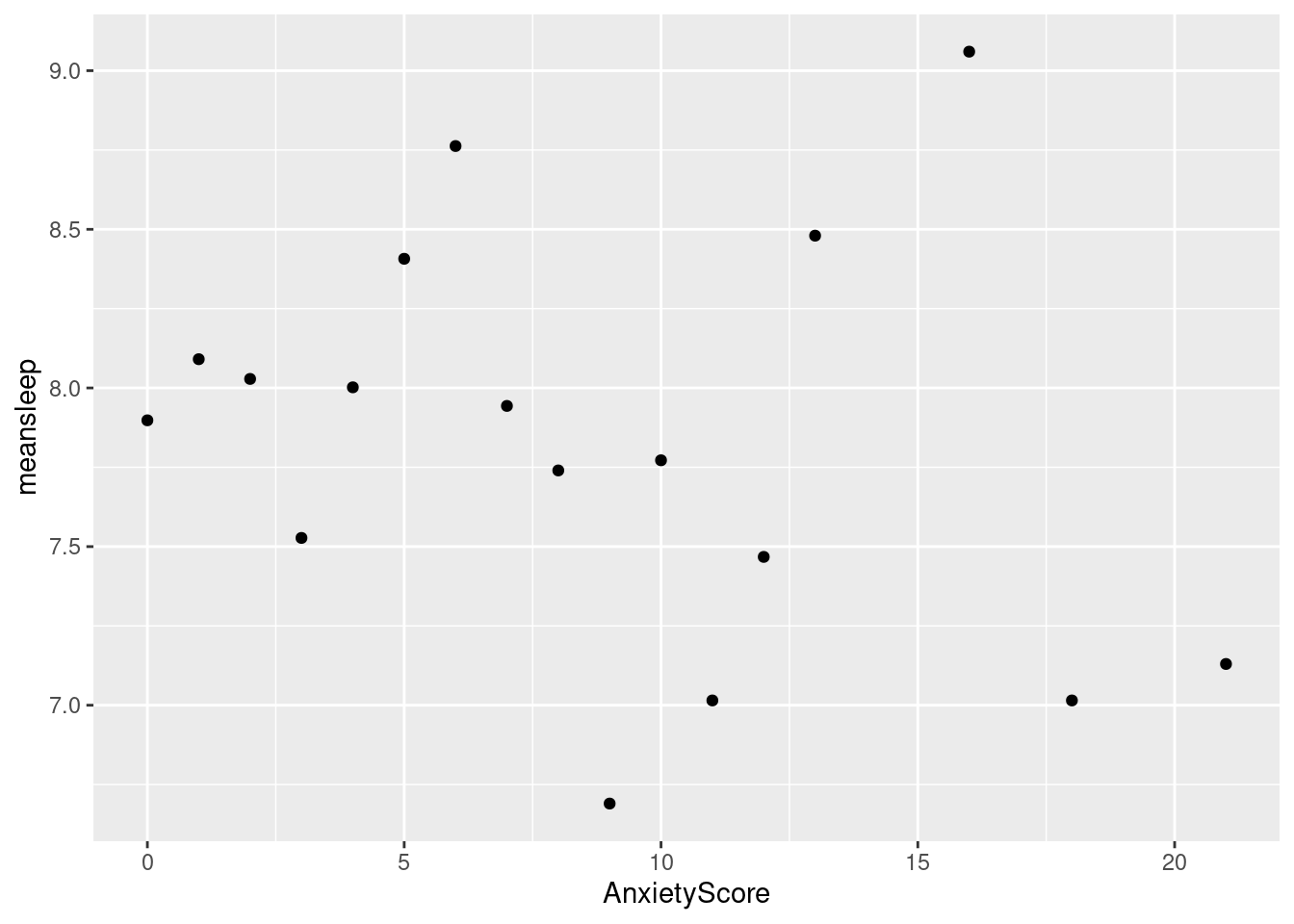
3.4 Types of visualizations
#Changing regions to a categorical variable.
happyplanet <- HappyPlanetIndex %>%
select(Country, Region, Happiness, LifeExpectancy, Footprint, GDPperCapita, Population)
happyplanet$region <- as.factor(ifelse(HappyPlanetIndex$Region < 2, 'Latin America', ifelse(HappyPlanetIndex$Region < 3, 'Western Nations', ifelse(HappyPlanetIndex$Region < 4, 'Middle East', ifelse(HappyPlanetIndex$Region < 5, 'Sub-Saharan Africa', ifelse(HappyPlanetIndex$Region < 6, 'South Asia', ifelse(HappyPlanetIndex$Region < 7, 'East Asia', 'former Communist countries')))))))
happyplanet$footprint <- as.factor(ifelse(happyplanet$Footprint > 6, "High", ifelse(happyplanet$Footprint < 3, "Medium", "Low")))
happyplanet$footprint <- factor(happyplanet$footprint, levels=c("Low", "Medium", "High"))
str(HappyPlanetIndex)## 'data.frame': 143 obs. of 11 variables:
## $ Country : Factor w/ 143 levels "Albania","Algeria",..: 1 2 3 4 5 6 7 8 9 10 ...
## $ Region : int 7 3 4 1 7 2 2 7 5 7 ...
## $ Happiness : num 5.5 5.6 4.3 7.1 5 7.9 7.8 5.3 5.3 5.8 ...
## $ LifeExpectancy: num 76.2 71.7 41.7 74.8 71.7 80.9 79.4 67.1 63.1 68.7 ...
## $ Footprint : num 2.2 1.7 0.9 2.5 1.4 7.8 5 2.2 0.6 3.9 ...
## $ HLY : num 41.7 40.1 17.8 53.4 36.1 63.7 61.9 35.4 33.1 40.1 ...
## $ HPI : num 47.9 51.2 26.8 59 48.3 ...
## $ HPIRank : int 54 40 130 15 48 102 57 85 31 104 ...
## $ GDPperCapita : int 5316 7062 2335 14280 4945 31794 33700 5016 2053 7918 ...
## $ HDI : num 0.801 0.733 0.446 0.869 0.775 0.962 0.948 0.746 0.547 0.804 ...
## $ Population : num 3.15 32.85 16.1 38.75 3.02 ...3.4.1 Line Plots
A line plot is useful for visualizing trends over time.
#First we will group the data by year and find the average life expectancy for each year.
by_year1 <- gapminder %>%
group_by(year) %>%
summarize(meanlife = mean(lifeExp))
#In a line graph, we can graph how average life expectancy has increased over the years.
ggplot(by_year1, aes(x = year, y = meanlife)) +
geom_line(color = "pink") + expand_limits(y = 0) + ylab("Average Life Expectancy")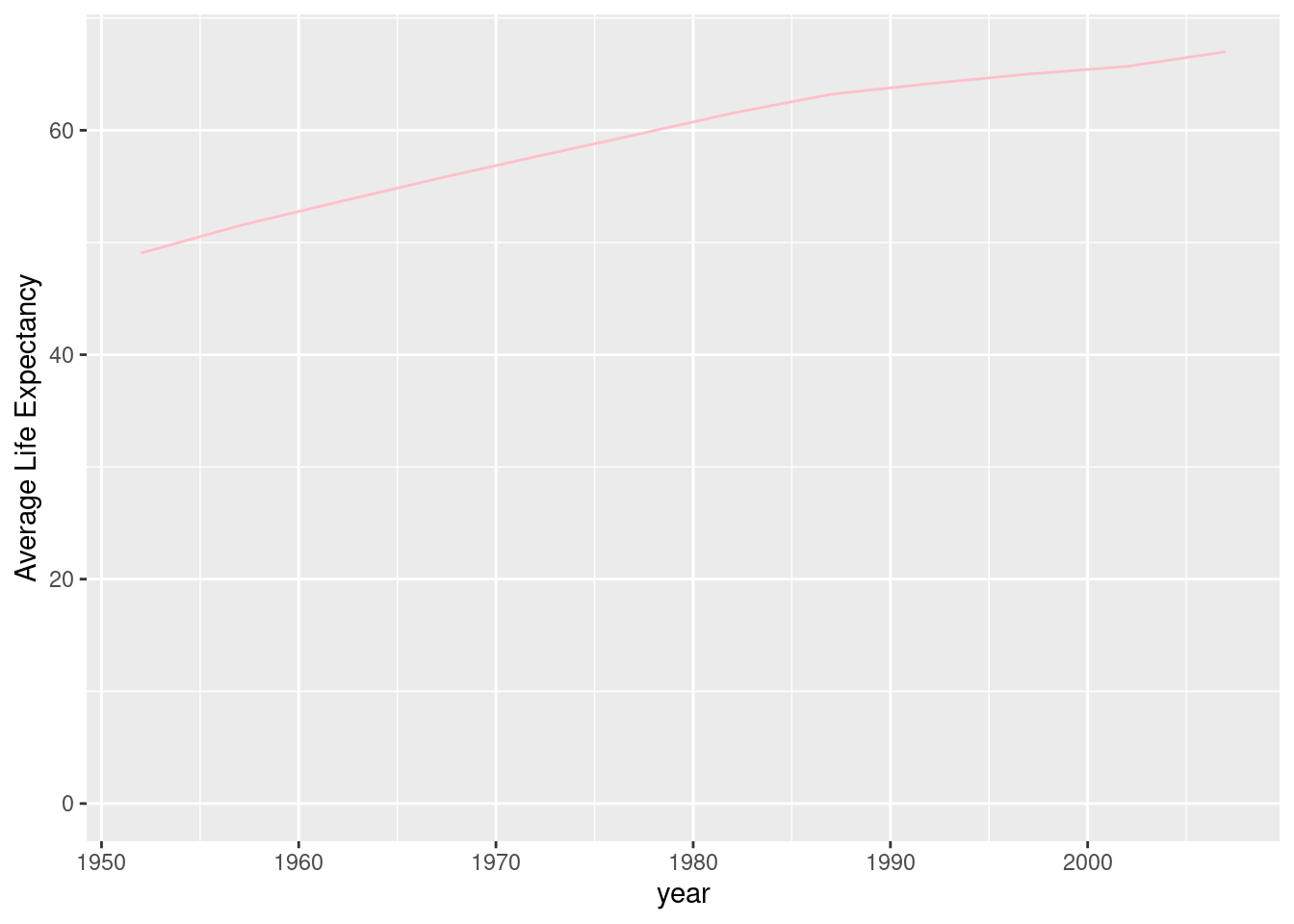
#Here we will group by both year and continent to find the average life expectancy for each continent in each year.
by_year_cont <- gapminder %>%
group_by(year, continent) %>%
summarize(meanLifeExp = mean(lifeExp))## `summarise()` has grouped output by 'year'. You can override using the
## `.groups` argument.#We can now graph our new data set and use color to differentiate between continents.
ggplot(by_year_cont, aes(x = year, y = meanLifeExp, color = continent)) +
geom_line() + expand_limits(y = 0) + ylab("Average Life Expectancy")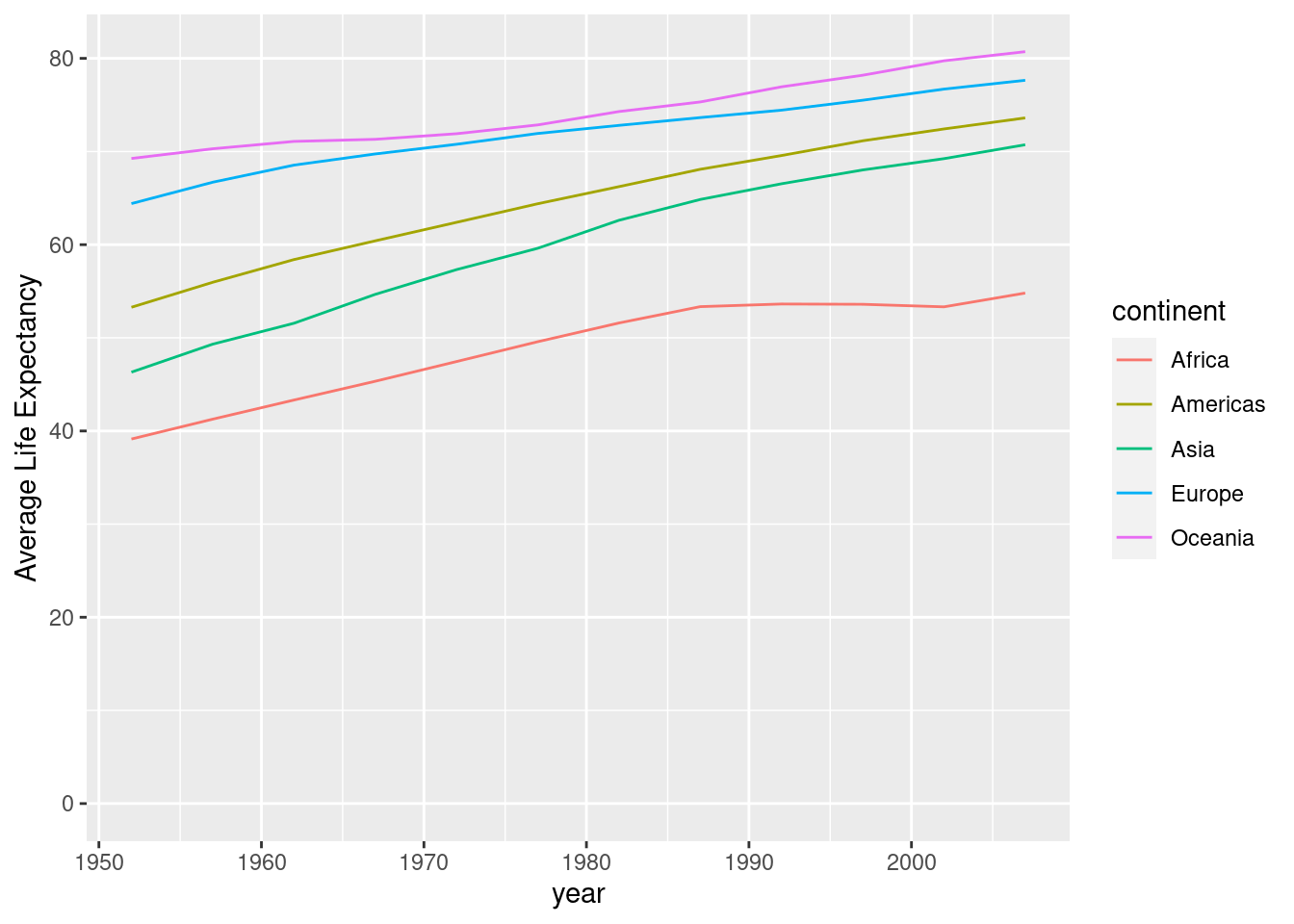
3.4.2 Bar Plots
A bar plot is useful for visualizing summary statistics.
#First we group the data set by region and find the mean happiness score within each region.
by_region <- happyplanet %>%
group_by(region) %>%
summarize(meanhappy = mean(Happiness))
#Then we plot the new data set in a bar plot.
ggplot(by_region, aes(x = region, y = meanhappy)) +
geom_col(fill = "pink", color = "white") + ylab("Mean Happiness Level")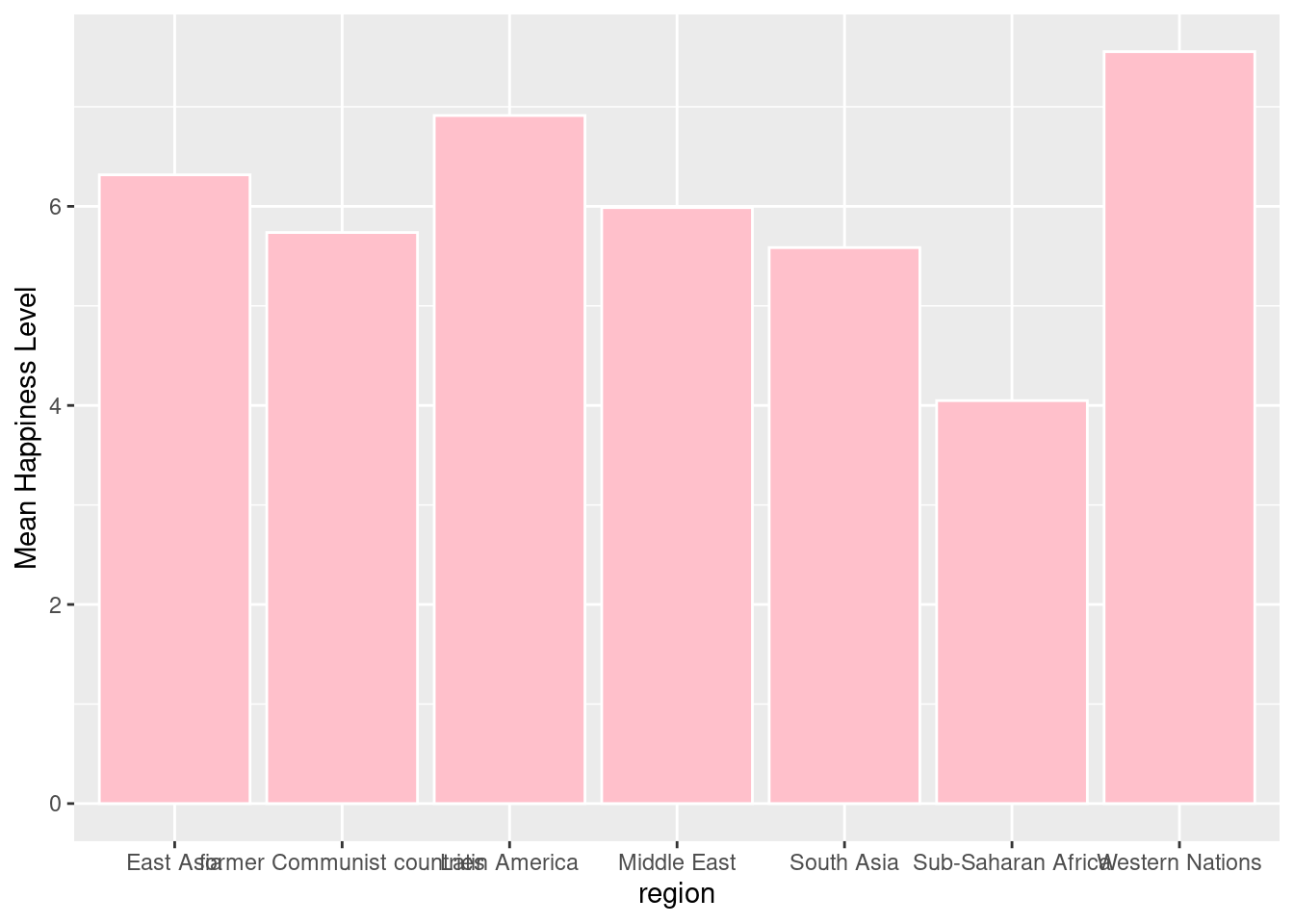
#We can also use a bar plot to see the differences in happiness level between countries in a specific region. Here we can filter the data to only include countries from South Asia.
SA_happiness <- happyplanet %>%
filter(region == "South Asia")
#Using a bar plot, we can visualize the happiness levels between these South Asian countries.
ggplot(SA_happiness, aes(x = Country, y = Happiness)) +
geom_col(fill = "pink", color = "white")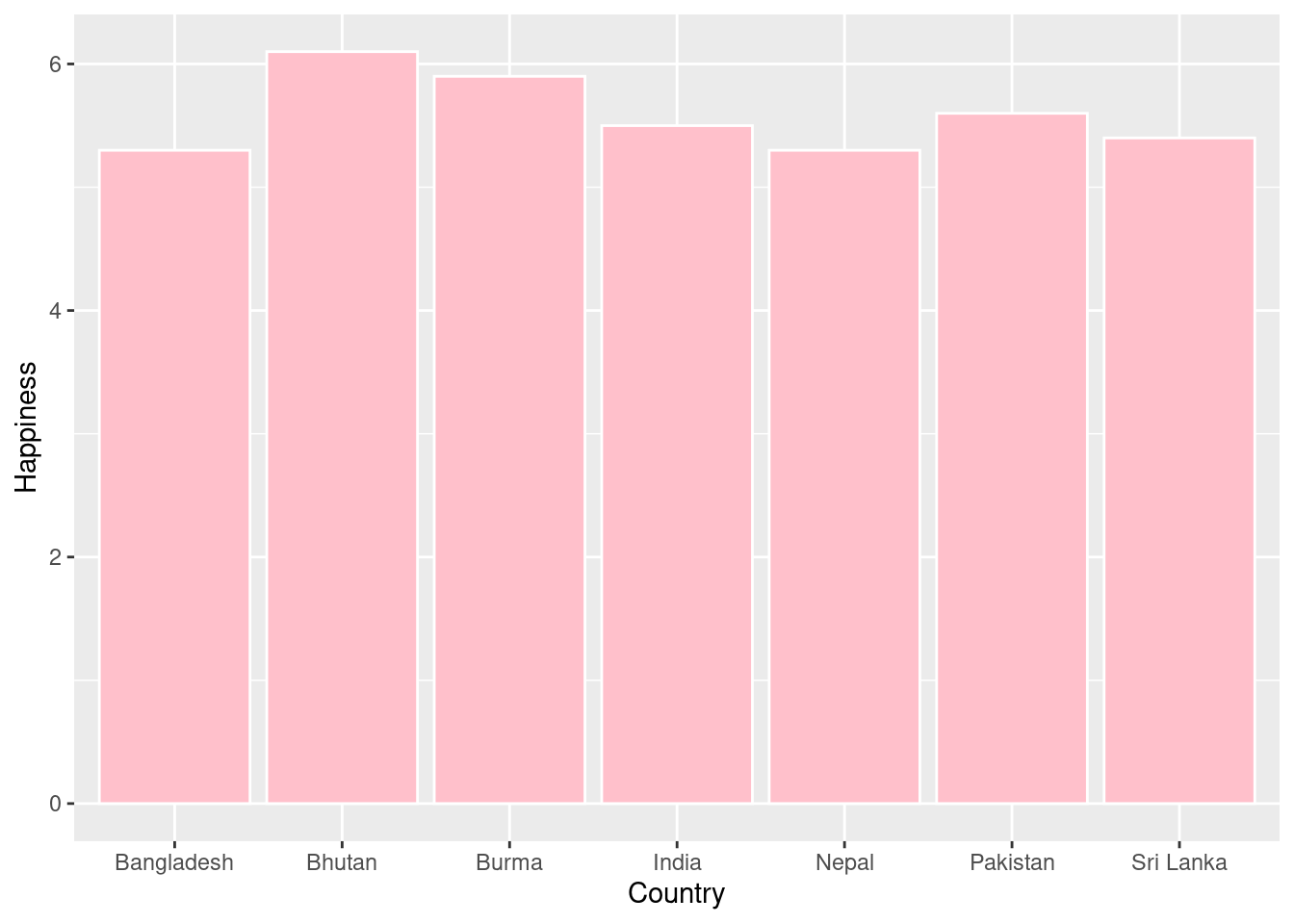
3.4.3 Histograms
A histogram is useful for examining the distribution of a numeric variable.
#Here we can see how common each happiness score is. It appears the most common score is around 5.5.
ggplot(happyplanet, aes(x = Happiness)) +
geom_histogram(binwidth = .5, color = "white", fill = "pink")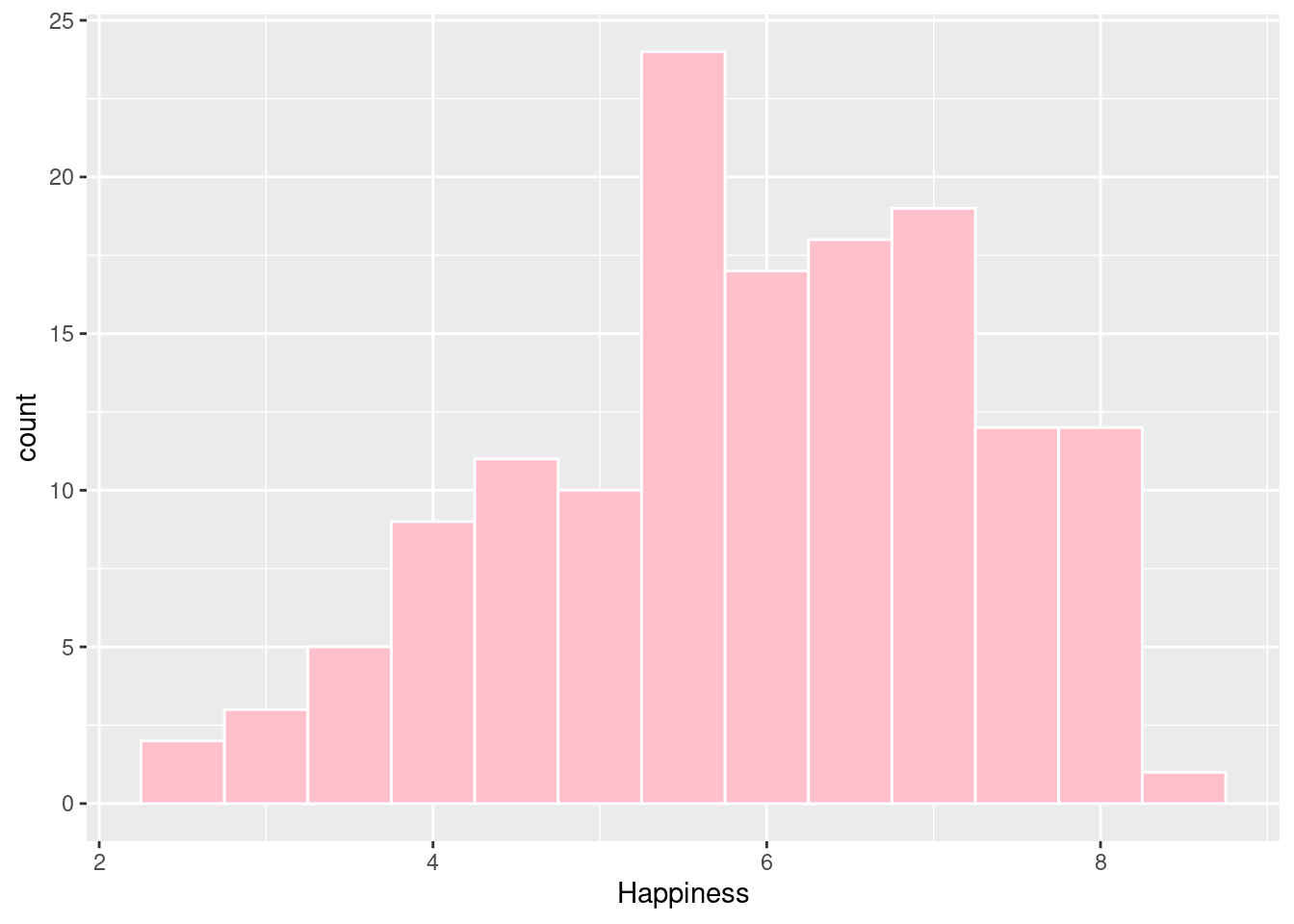
#First we will change population from in millions to the actual population.
happyplanet <- happyplanet %>%
mutate(population = Population * 1000000)
#Creating a histogram of population without using a log scale.
ggplot(happyplanet, aes(x = population)) +
geom_histogram(bins = 20, color = "white", fill = "pink")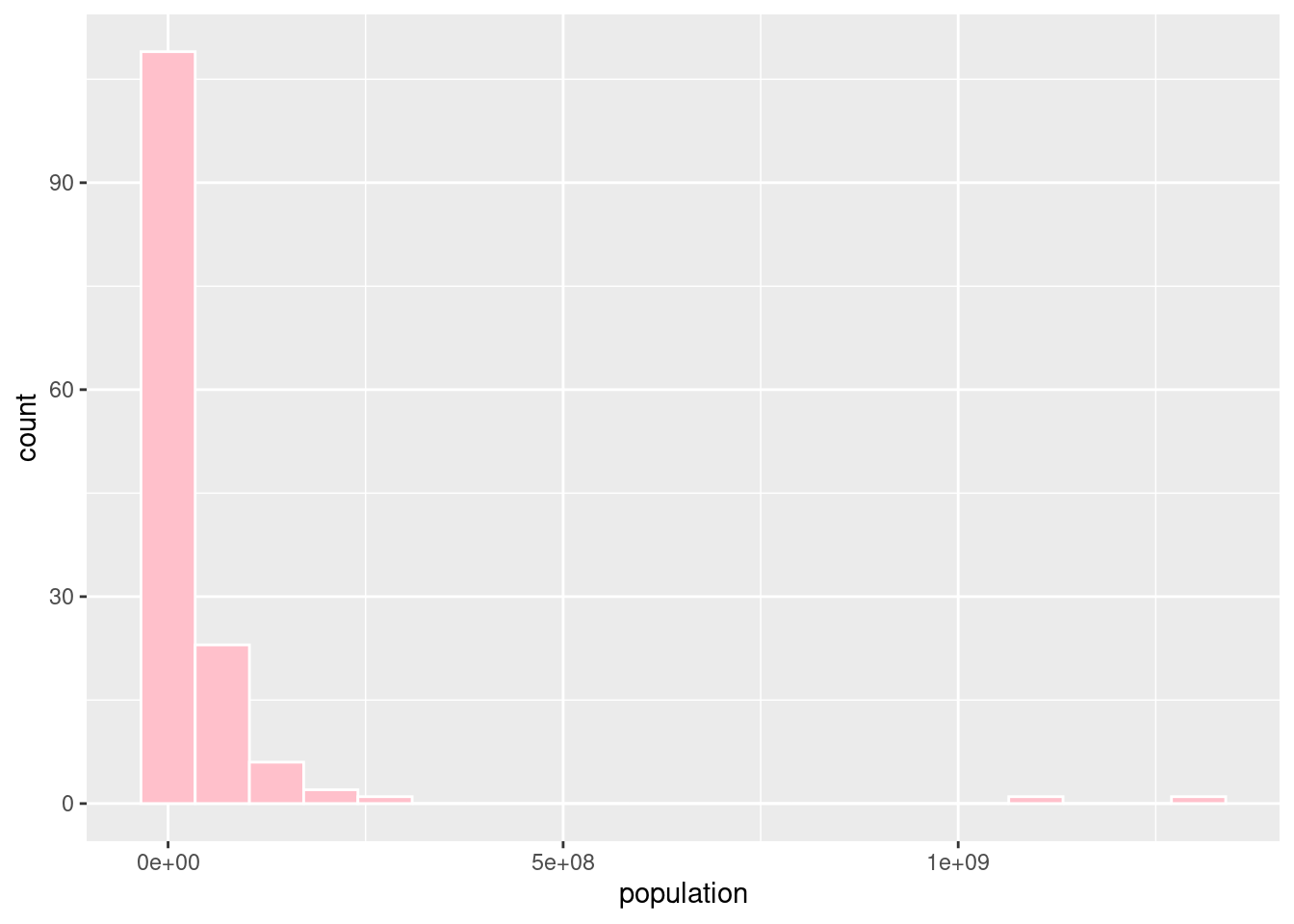
#Creating a histogram of population using a log scale
ggplot(happyplanet, aes(x = population)) +
geom_histogram(bins = 20, color = "white", fill = "pink") + scale_x_log10()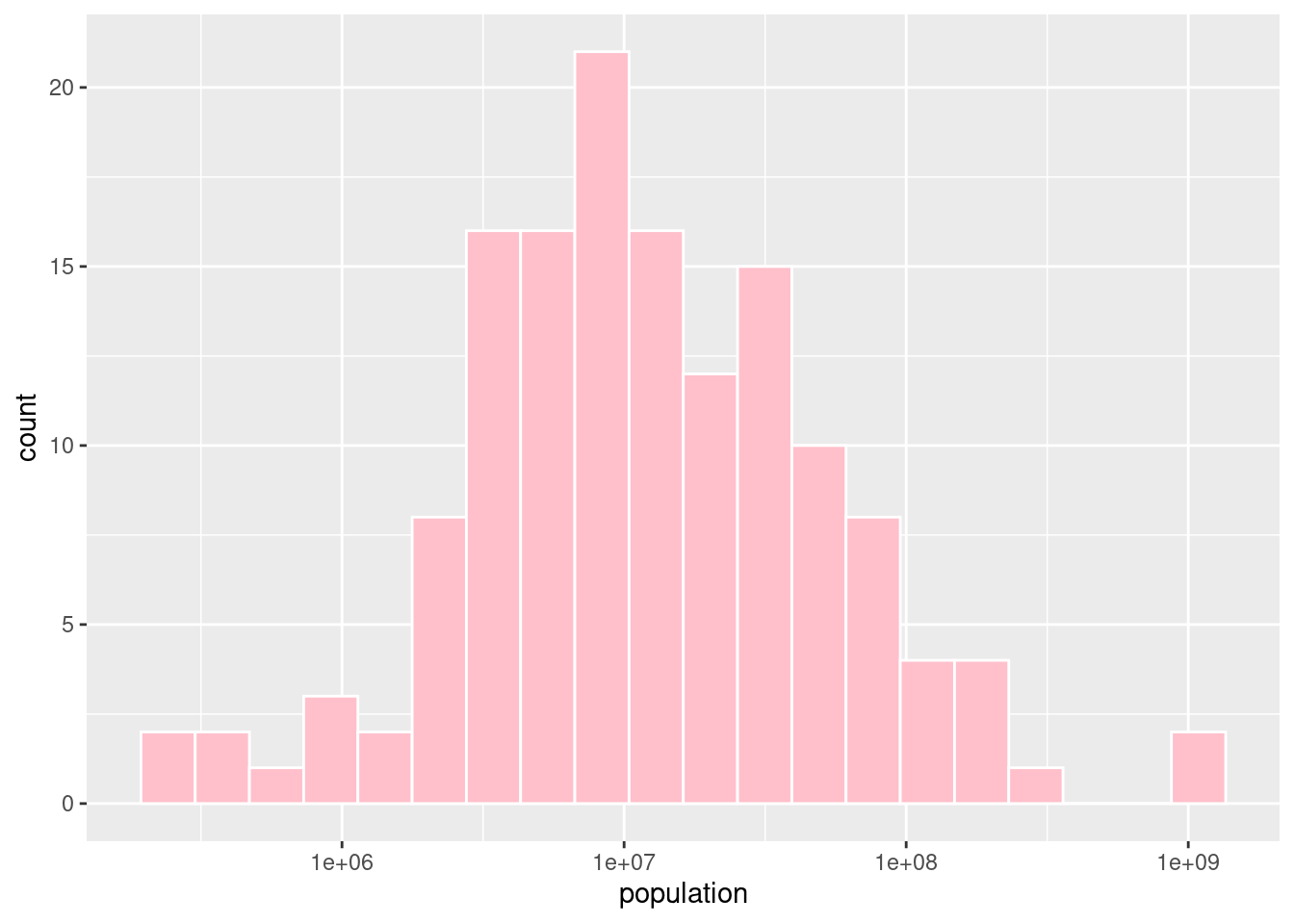
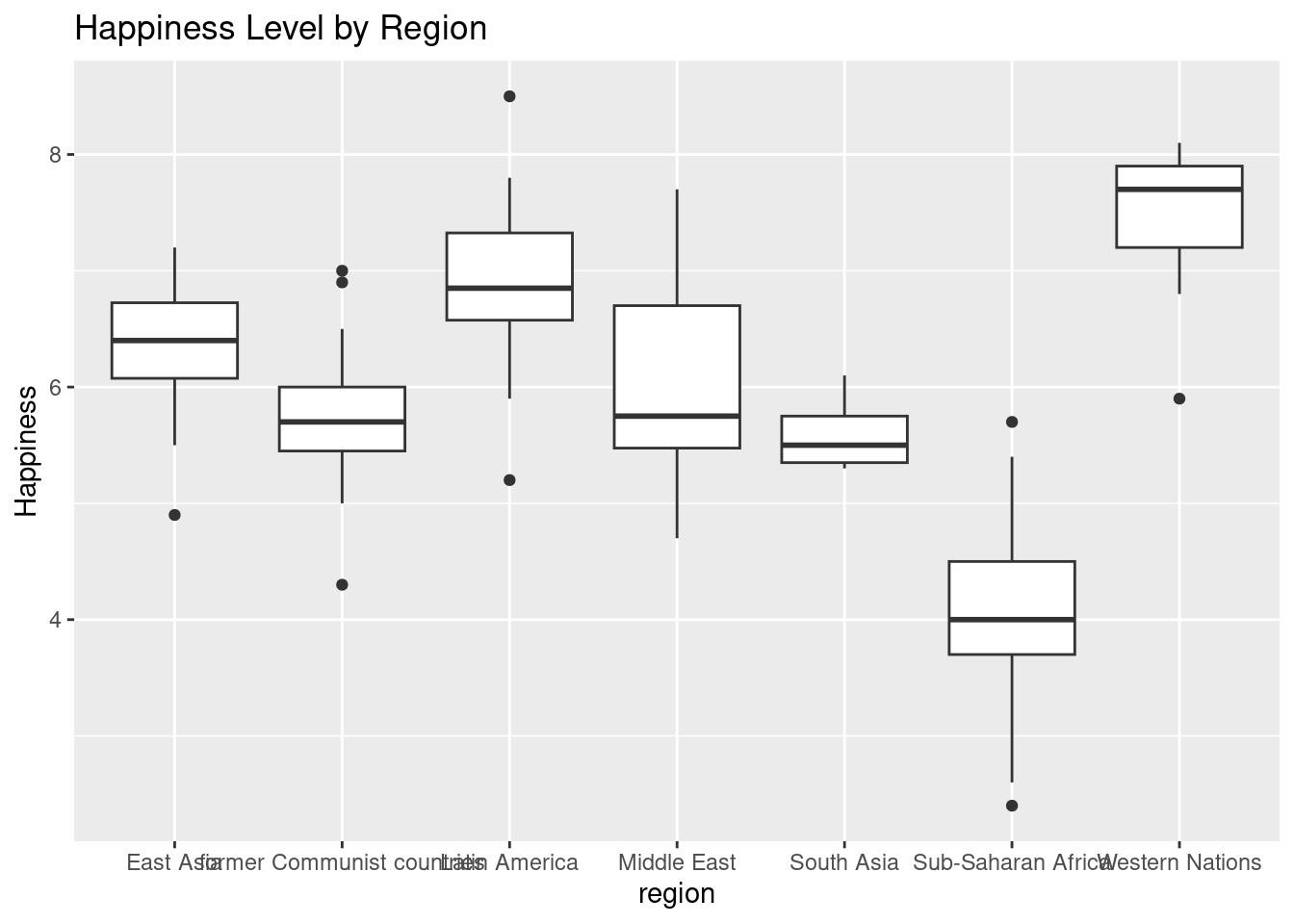
3.4.4 Boxplots
A boxplot is useful for comparing a distribution of values across several groups.
#Box plot that maps happiness levels for each region. We can also add titles to our graphs using labs(title = "").
ggplot(happyplanet, aes(x = region, y = Happiness)) +
geom_boxplot() + labs(title = "Happiness Level by Region")